
21
PLoS Genet. 2012;8(7):e1002854.
Genetic variants in REC8, RNF212, and PRDM9
infuence male recombination in cattle.
Sandor C, Li W, Coppieters W, Druet T, Charlier C, Georges M.
Abstract
We use >250,000 cross-over events identifed in >10,000 bovine sperm cells to perform an
extensive characterization of meiotic recombination in male cattle. We map Quantitative
Trait Loci (QTL) infuencing genome-wide recombination rate, genome-wide hotspot usage,
and locus-specifc recombination rate. We fne-map three QTL and present strong evidence
that genetic variants in REC8 and RNF212 infuence genome-wide recombination rate, while
genetic variants in PRDM9 infuence genome-wide hotspot usage.
PLoS Genet. 2012;8(3):e1002581
.
A splice site variant in the bovine RNF11 gene
compromises growth and regulation of the
infammatory response.
Sartelet A, Druet T, Michaux C, Fasquelle C, Géron S, Tamma N, Zhang Z, Coppieters W,
Georges M, Charlier C.
Abstract
We report association mapping of a locus on bovine chromosome 3 that underlies a Mende-
lian form of stunted growth in Belgian Blue Cattle (BBC). By resequencing positional candi-
dates, we identify the causative c124-2A>G splice variant in intron 1 of the RNF11 gene, for
which all afected animals are homozygous. We make the remarkable observation that 26% of
healthy Belgian Blue animals carry the corresponding variant. We demonstrate in a prospec-
tive study design that approximately one third of homozygous mutants die prematurely with
major infammatory lesions, hence explaining the rarity of growth-stunted animals despite the
high frequency of carriers. We provide preliminary evidence that heterozygous advantage
for an as of yet unidentifed phenotype may have caused a selective sweep accounting for
the high frequency of the RNF11 c124-2A>G mutation in Belgian Blue Cattle.
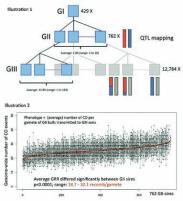
I
llustration 1 : Three-generational pedi-
grees used to map genetic determinants
of variation in male recombination rate
in cattle. Each pedigree is composed
of one grand-sire (GI generation) with
1.84 GII sons on average (range: 1 to
20). Each GII sire has 12.85 GIII sons on
average (range: 1 to 163). We have used
the available SNP genotypes to identify
259,752 crossing-over events that oc-
curred in the sperm cells transmitted by
the GII sires to their GIII sons. The two
homologues of a GII bull chromosome
are depicted in red and blue, recombina-
tion events in its GIII sons are highlighted
by switch between red and blue color.
Illustration 2: Variation of the Global Recombination Rate (GRR) in GII sires. Genome-wide numbers
of crossing-over events (CO) are measured in GIII sons. Black dots correspond to the total number of
crossing-over events identifed in the paternal genome of 10,192 GIII sons sorted by GII sire. The red dots
mark the GRR for each GII sire.