

58
Bioinformatics
The GIGA-Bioinformatics platform participates to the analysis of biomedical data generated by all the
other platforms through data analysis services and the development of new softwares and algorithms.
Working in close collaboration with the scientists of the Bioinformatics and Modeling Research unit, the
Bioinformatics platform activally participates in studies involving genome sequencing, clinical or trans-
criptomics data or proteomic profles. In addition, the high-performance computing infrastructure of the
platform delivered tens of thousands CPU days for data analyses and storage.
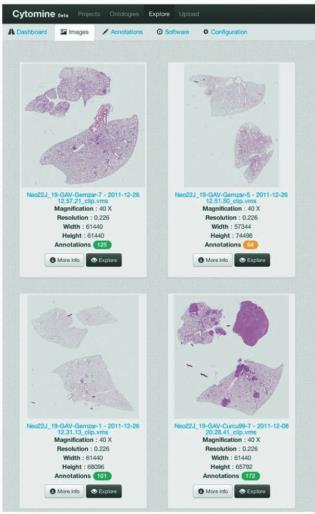
Moreover, the platform has been involved in the CYTOMINE software development (www.cytomine.
be), a rich internet application through which the scientist could
- Upload images in various formats (TIFF, JPEG, PNG, JPEG2000, Aperio SVS, Hamamatsu VMS, 3DHistech
MRXS, Leica SCN, ...)
- Organize the images into «picture folders » corresponding to a specifc study or experiment, with spe-
cifc permission rights
- Explore high-resolution (Gigapixel) images at multiple resolutions using modern navigation interfaces
inspired by web-based geographic applications such as Google Maps and OpenStreetMap
- Annotate the images by drawing multiple regions of interest and associate them to user-defned terms
from structured vocabularies (ontologies). Take advantage of our image retrieval algorithms to speed up
and consolidate annotations, hence facilitating the shaping of pathology atlases
- Share and Review images and their annotations with collaborators worldwide, discuss cases with your
colleagues and follow other user’s observation paths in real-time
- Analyze sets of images with fast image processing and generic machine learning methodologies imple-
mented on modern, multi-core architectures and computing clusters. Tissue and cell type recognition
algorithms can be trained by experts to detect and describe the most relevant biological «objects».
For computer scientists, we also ofer the possibility to plug their own algorithms using a RESTful
standardized programming interface that can be implemented in Java/Python/... clients.
In 2012 the software has been used by 30 biomedical researchers, and it hosted more than 8000 images
and 50 000 annotations related to studies in cancer, infammation, and toxicology.
Contact
Raphaël Marée
bioinformatics.giga@ulg.ac.be
www.giga.ulg.ac.be/bioinformatics